Table of Contents
Refine Queries with Metadata
Through the OpenBiomechanics Project, Driveline Baseball has made elite-level motion capture data publicly available. The data can be downloaded from Driveline's GitHub repository.
Having access to large datasets is a great asset, but processing these large datasets can take time (and patience). Sift is designed to streamline this process. The steps in this tutorial describe how to create CMZ files from the raw C3D files from pitching and batting data sets, then inspect and group the data in Sift.
Downloads and Relevant Links
Begin by downloading the following two zip files.
- OpenBiomechanics Project: Driveline Baseball data (download the entire biomechanics project with sub folders)
Please note, these downloads contain a large amount of data. Building CMZs is a slow process, so if you want to speed it up to follow along with building CMZ portion of this tutorial cut the number of datasets under openbiomechanics-main down to 3-5 subjects.
If you want to skip the Build CMZ step of this tutorial and focus on the metadata applications, you can:
- download this pre-built CMZ workspace and skip to Refining Hitting Data
- skip to the Build Your Own Metadata File example.
For more technical information about loading metadata, see: Metadata Documentation
Build CMZs for Hitting Data
First open a new Sift workspace.
On the Load Page, select “Load C3Ds”
In the Load C3Ds Dialog under Generic Builder:
- Set the path to: C:\…\openbiomechanics-main\baseball_hitting\data\c3d
- Set the Metadata File to : C:\…\openbiomechanics-main\baseball_hitting\data\metadata.csv
- Set the MDH File to: C:\…\openbiomechanics-main\baseball_hitting\code\v3d\model\hitting_v1_model_hybrid.mdh
- Add the following scripts and ensure they are in the correct order:
- C:\…\CMotion Files\Filter.v3s
- C:\…\CMotion Files\Events.v3s
- C:\…\openbiomechanics-main\baseball_hitting\code\v3d\CMO.v3s
- Click Create CMZs
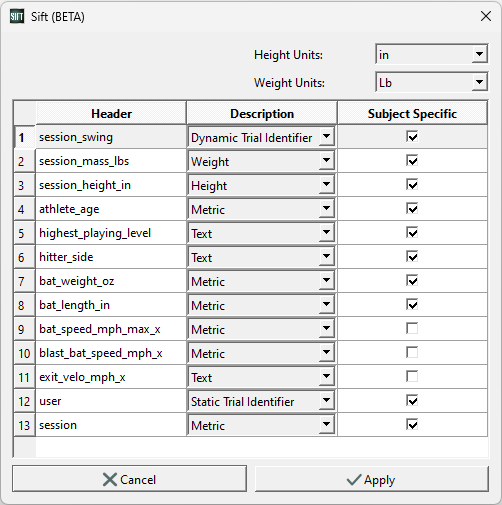
- A metadata dialog box will appear after clicking Create CMZs. Make the following changes in this box:
- Height Units: in
- Weight Units: Lb
- session_swing: Dynamic Trial Identifier
- user: Static Trial Identifier
- Check “Subject Specific” next to highest_playing_level, hitter_side, bat weight/length, and Click “Apply”.
- CMZ files may take a while to build. Check the status bar in the bottom left hand corner of the interface for the build status.
- After the files are built, check that the Library Path dialog is set correctly and hit “Load” in the Load Library dialog.
- On the Load Page, you will see all of the workspaces for the hitting data have been created and loaded.
Refining Hitting Data
From here on it is assumed that you either have: a) built CMZs from scratch using the hitting data set, or b) downloaded the pre-made CMZs linked above.
The zip file openbiomechanics-main has a metadata file for the hitting data set which you can view at your convenience. This metadata file has a number of categories including player age, bat weight, hitting side etc.
Let's say we want to query this data set, but we only want data from subjects that are under age 20.
We start by loading library with the hitting data set, navigating to the Query Builder dialog, and opening the Auto-Populate dialog.
Under “Combine Sides”, select Baseball from the drop down. Here you will see the info box auto-populated by the meta data in the CMZ files.
Additional refinements, under the “Refinements” tab, find the “Refine using signal” checkbox and click “Add Signal”. Then you can enter your query refinements for athletes under age 20:
When refining queries it can be helpful to have the metadata file open, so that you can see the values it contains. Note that in this case, accessing athlete_age has been set as a global metric, and you will not be able to find the meta folder unless Global Value is checked.
The result of this procedure is that data is queried only if it meets this refinement category, and the refinement name will be applied as a suffix to the end of the signal name.
When you hit “Create” on the Auto-Populate Dialog, you will see the queries appear for athletes under 20, and with L and R batting sides combined for each query.

Build Your Own Metadata File
Let's say you have three subjects worth of data where each subject walks at four different speeds and has a different weight.
In this example let us say we want to include metadata on the subject's walking speed and weight. To create a .csv file for this metadata, start by creating a row of headers. The minimum requirements for a metadata file include the static file name and dynamic file name. After that, we can add the other parameters we are interested in.
In the example below, we can see that there are four headers (static_model, dynamic_file, walking_speed and subject_mass).
Here you can edit the description fields and subject specific fields. After you are satisfied with the fields, click “Apply” to build CMZs and load the library.
Using Metadata to Refine Queries
Once your data is loaded it can be queried. This example shows how you can use metadata to refine queries.
In this instance, we have data where three subjects walk at three different speeds.
If you navigate from the Query Builder dialog to the Auto-Populate menu, In the Signals tab Select Metric as the signal type and meta as the signal folder. Now switch over to the Refinements tab, click  Add Signal, create three refinements as seen in the picture below, and then hit
Add Signal, create three refinements as seen in the picture below, and then hit  Create.
Create.
Since I want to refine my data by walking speed in this example, I name the refinement “2.0”, choose the data under METRIC>meta>walking_speed>X and indicate that the value must be “Equal to” 2.0. I can then save my refinements as an .xml file which can be loaded at any time if I want to re-query my library.