Table of Contents
Looking at Large Public Data Sets
Biomechanics is coming into a new age, where large datasets are the norm and individually processing the files can be tiresome and time consuming. Captured over long periods of time, datasets can be inconsistent, which makes it difficult to automate working with them.
In this tutorial, we will be going through common issues seen while working with large public biomechanics datasets, through the use of a large dataset released which tracks the full body gait of both stroke survivors and able-bodied participants.
Large Datasets
The dataset used for this tutorial is the raw motion data collected by (Van Criekinge et al [1]), which includes the full-body motion capture gait of 138 able-bodied adult participants and 50 stroke survivors. They include Full Body Kinematics (PiG Model), Kinetics, and EMG Data across a wide variety of ages, heights and weights. This is an open sourced dataset, which allows for free and open science, supports the growing bio-mechanical research community and increases the ability for scientists to duplicate results of tests.
This dataset is unique in its size and quality for an open source dataset. Large datasets are essential for scientific observations to be able to accurately draw conclusions, and are becoming the norm within the bio-mechanical community. Because of their nature as large collections of data, there are often unique problems relating to working with such datasets, which is to be explored here. Datasets such as these can be generated over a long period of time, where people, standards or equipment may change. Large datasets can also be the culmination of several different projects, which further leads to inconsistencies. The most frequent issues we found were due to inconsistencies within the measured dataset: it can be difficult to keep a rigid structure to any large dataset, but inconsistencies can wreck the ability to automate tasks, so we’ll show how you can effectively process the data in large datasets like this.
Dataset
The dataset used for able-bodied participants is available here, while the dataset used for stroke survivors is available here (also contains SubjectChar.xlsx, an excel file with survivors parameters).
The authors have made an already-processed dataset available for use, but for the purposes of this tutorial, we shall be using the original, raw data.
Please find all of the needed pipeline and model files here.
File Naming Conventions
A common inconsistency in large datasets can be the usage of filenames. Filenames should convey information about a file in a manner that can be easily referenced, eg: calibration files should be named differently from motion files so that they can be separated, and motion files should have a trial number in their file name (if applicable), etc.
In this dataset, we found inconsistencies within filenames, as well as an error relating to the file name. Particularly within the stroke survivors, there was no consistent naming convention across all of the motion files, other than all including the letters “BWA”. How the trial number was conveyed was done in a variety of different methods, including separating with spaces, underscores or nothing, as well as prepending a “0” to the trial number. Ultimately this forces the user to use wildcards to collect all the motion files at once, and limits their ability to individually select motion files.
There are several files which need to be altered or deleted:
- Stroke Survivor 47 (TVC47) has a mislabeled calibration file. This must be renamed from “BWA (0).c3d” to something following their calibration conventions, such as “TVC-47 Cal 0.c3d”.
- Stroke Survivor 38 (TVC38) does not have a valid calibration file. The corresponding folder should be deleted (TVC38).
- Able-Bodied Participants 37 (SUBJ37), 42 (SUBJ42), and 118-138 (SUBJ118-SUBJ138) follow a different model to mark the pelvis, or are missing necessary data. The corresponding folders should be deleted (SUBJ37, SUBJ42, SUBJ118-SUBJ138)
Building Models
This data set primarily used the Plug in Gait marker set, however, unfortunately not all the standard Plug in Gait markers were used. For example the right and left upper arm markers were missing, so only a model of the lower body can be made, see the tutorial here to walk through the process of making a lower body plug in gait model.
The subset of able-bodied participants used in this tutorial has three markers for the pelvis as opposed to the four used for the stroke survivors; two separate models need to be made. The default values provided in the tutorial can be used to build the models, as the subject data will be entered in via the processing pipeline.
For the Stroke Survivors, it is important to build the original model without any subject-prefixes (managed later in this tutorial). Participant 58 should be used for this, as their calibration file does not have any attached subject-prefixes in the raw data.
The respective models we used have been provided in this tutorials files: stroke_model.mdh and healthy_model.mdh.
Inconsistent Subject Prefixes (Stroke Survivors)
Most of the markers within the static files provided are prefixed with the subject ID, however the dynamic file markers are not, this will prevent the built model from linking with the dynamic markers. To begin, we will ensure that all static files include a subject-prefix, and then we will prepend these to the markers within the dynamic files.
Static file marker:

Dynamic file marker:
Stroke Survivors 47 and 58 (TVC47 and TVC58) are both missing subject prefixes. To rectify, run the pipeline file add_calibration_prefixes.v3s once for each of these stroke survivors, changing the “/FOLDER” section of the pipeline parameter “Set_Pipeline_Parameter_To_List_Of_Files” to the folder containing each stroke survivors files (TVC47 and TVC58).
The pipeline “convert_subject_prefix_stroke.v3s” can be used to prepend the subject IDs to all the marker points in the dynamic files. The subject ID is stored in the PARAMETERS::SUBJECTS::NAMES signal within the dynamic files, however they use a dash instead of an underscore, “BWA-2” vs “BWA_2” these still wont match. String expressions can be used to convert the subject ID strings to match the static files.
Run the pipeline convert_subject_prefix_stroke.v3s, and select the folder containing the stroke survivors (50_StrokePiG) when prompted.
Below is an explanation of the important sections of the file “convert_subject_prefix_stroke.v3s”.
Set a pipeline parameter to a list of all dynamic files contained in the “50_Stroke_PiG”:
Set_Pipeline_Parameter_To_Folder_Path /PARAMETER_NAME=FOLDER ! /PARAMETER_VALUE=VISUAL3D_DEFAULT_DATA_FOLDER ! /FROM_MOTION_FILE_IN_WORKSPACE= ! /PARAMETER_VALUE_SEARCH_FOR= ! /PARAMETER_VALUE_REPLACE_WITH= ! /PARAMETER_VALUE_APPEND= ! /SET_PROMPT=Select a directory ! /ALPHABETIZE=TRUE ; Set_Pipeline_Parameter_To_List_Of_Files /PARAMETER_NAME=DYNAMIC_FILES /FOLDER=::FOLDER /SEARCH_SUBFOLDERS=TRUE /FILE_MASK=*BWA*.c3d /ALPHABETIZE=FALSE ! /RETURN_FOLDER_NAMES=FALSE ;
Loop through all dynamic files:
For_Each /ITERATION_PARAMETER_NAME=DYNAMIC_FILE ! /ITERATION_PARAMETER_COUNT_NAME= /ITEMS=::DYNAMIC_FILES ;
Open the current file:
File_Open FILE_NAME=::DYNAMIC_FILE ! /FILE_PATH= ! /SUFFIX= ! /SET_PROMPT=File_Open ! /ON_FILE_NOT_FOUND=PROMPT ! /FILE_TYPES_ON_PROMPT= ;
Get the Participant ID from PARAMETERS:SUBJECT:NAMES
Set_Pipeline_Parameter_To_Data_Value /SIGNAL_TYPES=PARAMETERS /SIGNAL_FOLDER=SUBJECTS /SIGNAL_NAMES=NAMES ! /SIGNAL_COMPONENTS=ALL_COMPONENTS /PARAMETER_NAME=SUBJECT_ID ;
Get the preceding half of the Participant ID using the STRING_LEFT expression, since all IDs begin with BWA we can hard code the index value:
Set_Pipeline_Parameter_From_Expression
PARAMETER_NAME=LEFT_SUBJECT_ID
EXPRESSION=STRING_LEFT("&::SUBJECT_ID&",3)
/AS_INTEGER=FALSE
;
Get the length of the participant id using the STRING_LENGTH expression:
Set_Pipeline_Parameter_From_Expression
/PARAMETER_NAME=SUBJECT_ID_LENGTH
/EXPRESSION=STRING_LENGTH("&::SUBJECT_ID&")
! /AS_INTEGER=TRUE
;
Get the index of the dash using the STRING_FIND expression
Set_Pipeline_Parameter_From_Expression
/PARAMETER_NAME=DASH_INDEX
/EXPRESSION=STRING_FIND("&::SUBJECT_ID&","-",0)
! /AS_INTEGER=TRUE
;
Subtract the DASH_INDEX from SUBJECT_ID_LENGTH to get the negative index of the dash, subtract that value by one to get the value needed for STRING_RIGHT (the negative index of the character immediately following the dash):
Set_Pipeline_Parameter_From_Expression
PARAMETER_NAME=RIGHT_SUBJECT_ID
/EXPRESSION=STRING_RIGHT("&::SUBJECT_ID&",&::DASH_NEGATIVE_INDEX&)
!/AS_INTEGER=FALSE
;
Get the characters in the subject id following the dash using STRING_RIGHT:
Set_Pipeline_Parameter_From_Expression
/PARAMETER_NAME=RIGHT_SUBJECT_ID
/EXPRESSION=STRING_RIGHT("&::SUBJECT_ID&",&::DASH_NEGATIVE_INDEX&)
!/AS_INTEGER=FALSE
;
Concatenate LEFT_SUBJECT_ID and RIGHT_SUBJECT_ID with an underscore between them:
Set_Pipeline_Parameter /PARAMETER_NAME=SUBJECT_ID_CONVERTED /PARAMETER_VALUE=_ ! /PARAMETER_VALUE_SEARCH_FOR= ! /PARAMETER_VALUE_REPLACE_WITH= /PARAMETER_VALUE_PREFIX=::LEFT_SUBJECT_ID /PARAMETER_VALUE_APPEND=::RIGHT_SUBJECT_ID ! /MULTI_PASS=FALSE ;
Get a list of all target names
Set_Pipeline_Parameter_To_List_Of_Signal_Names /PARAMETER_NAME=TARGETS /SIGNAL_TYPES=TARGET /SIGNAL_FOLDER=ORIGINAL ;
Using the updated subject ID and the list of target names, use the modify participants parameters pipeline function the add the needed subject prefixes, ensuring OVERWRITE_C3D_FILE is set to true so the changes are preserved:
Modify_C3D_Subjects_Parameters ! /INCLUDE_CAL_FILE=TRUE /IS_STATIC=0 /SUBJECTS_USES_PREFIXES=1 ! /REMOVE_EXISTING_LABEL_PREFIXES=TRUE ! /REMOVE_EXISTING_PREFIX_PARAMETERS=FALSE /SUBJECTS_LABEL_PREFIXES=::SUBJECT_ID_CONVERTED /PREFIX_MARKERS=::TARGETS ! /PREFIX_ROTATIONS= ! /PREFIX_C3D_PARAMETERS= ! /C3D_PARAMETER_GROUPS=SUBJECTS ! /MARKER_SETS= /SUBJECTS_NAMES=::SUBJECT_ID_CONVERTED ! /SUBJECTS_DISPLAY_SETS= ! /SUBJECTS_MODELS= ! /SUBJECTS_MODEL_PARAMS= /OVERWRITE_C3D_FILE=1 ;
Clear the workspace to preserve memory, and then close the loop:
File_New ;
End the loop
End_For_Each /ITERATION_PARAMETER_NAME=DYNAMIC_FILE ;
Inconsistent Subject Prefixes (Able Bodied)
The markers in the static files of the able bodied participants do not have subject prefixes, however a random selection of the dynamic file markers do have prefixes. In this case it makes sense to strip the subject prefixes from the select files that have them instead of adding them to every other file. This can be done using the “remove_subject_prefixes_healthy.v3s” pipeline file.
Run the pipeline remove_subject_prefixes_healthy.v3s, and select the folder containing the able-bodied participants (138_HealthyPiG_10.05) when prompted.
Because the prefixes are being removed instead of added, the pipeline is much simpler. Below is the primary change to the pipeline:
Using the modify C3D subjects parameters pipeline function clear the participant prefixes from the files, set OVERWRITE_C3D_FILE to true to ensure the changes are preserved, keep all other parameters at their default value:
Modify_C3D_Subjects_Parameters ! /INCLUDE_CAL_FILE=TRUE ! /IS_STATIC= ! /SUBJECTS_USES_PREFIXES= ! /REMOVE_EXISTING_LABEL_PREFIXES=TRUE ! /REMOVE_EXISTING_PREFIX_PARAMETERS=FALSE ! /SUBJECTS_LABEL_PREFIXES= ! /PREFIX_MARKERS= ! /PREFIX_ROTATIONS= ! /PREFIX_C3D_PARAMETERS= ! /C3D_PARAMETER_GROUPS=SUBJECTS ! /MARKER_SETS= ! /SUBJECTS_NAMES= ! /SUBJECTS_DISPLAY_SETS= ! /SUBJECTS_MODELS= ! /SUBJECTS_MODEL_PARAMS= /OVERWRITE_C3D_FILE=TRUE ;
Processing C3Ds into CMZs (Stroke Survivors)
Instead of manually processing each C3D file, we can use another pipeline to automate the process:
Because the parameters needed to accurately calculate the models are stored within the dynamic files, they must be opened first and the data saved to pipeline parameters so it can then be applied to the model.
Edit the pipeline command “Apply_Model_Template” in the pipeline process_c3ds_stroke.v3s to point to the model file you have built (or been provided: stroke_model.mdh).
Run the pipeline process_c3ds_stroke.v3s, and select the folder containing the stroke survivors (50_StrokePiG) when prompted.
It should also be mentioned that this dataset does not have reliable enough force plate data to accurately create gait events through kinetic events, and as such a kinematic model is used to generate these. This is done using the Zeni Method 1a (foot position relative to pelvis) from this tutorial.
Set a pipeline parameter to the current folder path
Set_Pipeline_Parameter_To_Folder_Path /PARAMETER_NAME=FOLDER /PARAMETER_VALUE= ! /PARAMETER_VALUE_SEARCH_FOR= ! /PARAMETER_VALUE_REPLACE_WITH= ! /PARAMETER_VALUE_APPEND= ;
Get a list of all folders within the 50_StrokePiG folder:
Set_Pipeline_Parameter_To_List_Of_Files /PARAMETER_NAME=Folders /FOLDER=::FOLDER /SEARCH_SUBFOLDERS=TRUE /FILE_MASK=*.c3d ! /ALPHABETIZE=TRUE /RETURN_FOLDER_NAMES=TRUE ;
Loop through the folders:
For_Each /ITERATION_PARAMETER_NAME=sub_folder ! /ITERATION_PARAMETER_COUNT_NAME= /ITEMS=::Folders ;
Calibration files are prefixed with TVC, set a pipeline parameter to all C3D files that start with TVC (there should only be one in each folder):
Set_Pipeline_Parameter_To_List_Of_Files /PARAMETER_NAME=SUBFOLDER_CALIBRATION /FOLDER=::sub_folder ! /SEARCH_SUBFOLDERS=FALSE /FILE_MASK=TVC*.c3d ! /ALPHABETIZE=TRUE ! /RETURN_FOLDER_NAMES=FALSE ;
Start a new workspace:
File_New ;
Open all dynamic files within the current folder:
File_Open /FILE_NAME=::sub_folder&BWA*.c3d ! /FILE_PATH= ! /SUFFIX= ! /SET_PROMPT=File_Open /ON_FILE_NOT_FOUND=FAIL ! /FILE_TYPES_ON_PROMPT= ;
Set all dynamic files to active:
Select_Active_File /FILE_NAME=::sub_folder&BWA*.c3d ! /QUERY= ! /SUBJECT_TAGS=NO_SUBJECT ;
Save all the necessary metrics from PARAMETERS:PROCESSING into pipeline parameters:
Set_Pipeline_Parameter_To_Data_Value /SIGNAL_TYPES= PARAMETERS /SIGNAL_FOLDER= PROCESSING /SIGNAL_NAMES= Bodymass ! /SIGNAL_COMPONENTS=ALL_COMPONENTS /PARAMETER_NAME= mass ; ... see attached pipeline for full list of commands ...
Visual 3Ds expected unit of measurement is meters so the knee widths, ankle widths and height need to be converted from mm to m:
Set_Pipeline_Parameter_From_Expression /PARAMETER_NAME=height /EXPRESSION=&::height&/1000.0 /AS_INTEGER=False ; ... see attached pipeline for full list of commands ...
Get the subject ID from PARAMETERS:SUBJECTS and save it to a pipeline parameter:
Set_Pipeline_Parameter_To_Data_Value /SIGNAL_TYPES=PARAMETERS /SIGNAL_FOLDER=SUBJECTS /SIGNAL_NAMES=NAMES ! /SIGNAL_COMPONENTS=ALL_COMPONENTS /PARAMETER_NAME=SUB_NAME ;
Create a hybrid model using the calibration file:
Create_Hybrid_Model /CALIBRATION_FILE=::SUBFOLDER_CALIBRATION ! /RANGE=ALL_FRAMES ;
Apply the model template:
Apply_Model_Template /CALIBRATION_FILE=::SUBFOLDER_CALIBRATION /SUBJECT_PREFIX=::SUB_NAME&: /MODEL_TEMPLATE=!!! SET PATH TO STROKE MODEL TEMPLATE !!! ! /SET_PROMPT=Open model file ! /VIEW_BUILDMODEL_RESULTS=2 ! /MISSING_TARGET_MESSAGE=FALSE ;
Set all the necessary model metrics and recalculate the model:
Set_Model_Metric ! /CALIBRATION_FILE= /METRIC_NAME= Mass /METRIC_VALUE= ::mass ! /SUBJECT= ; ... See the attached pipeline for full list of commands ...
Assign the model to the dynamic files:
Assign_Model_File /CALIBRATION_FILE=::SUBFOLDER_CALIBRATION /MOTION_FILE_NAMES=::sub_folder&BWA*.c3d ! /REMOVE_EXISTING_ASSIGNMENTS=FALSE ;
Set all files to active:
Select_Active_File /FILE_NAME= ALL_FILES ! /QUERY= ! /SUBJECT_TAGS=NO_SUBJECT ;
Filter and Interpolate available data
...See attached pipeline for examples of computation commands ...
Calculate Gait Events through the use of Kinematic signals
...See attached pipeline for examples of computation commands ...
Run all desired computations and then save the workspace to a CMZ using the subject ID as the file name and end the loop
..See attached pipeline for examples of computation commands ... File_Save_As /FILE_NAME=::SUB_NAME /FOLDER=::sub_folder ! /SET_PROMPT=Save CMZ file as ! /SAVE_EMBEDDED_GRAPHICS=FALSE ! /CREATE_FOLDER_PATH=FALSE ; End_For_Each /ITERATION_PARAMETER_NAME= sub_folder ;
Processing C3Ds into CMZs (Able-Bodied Participants)
Since the naming convention for the able-bodied participants is different from the stroke survivors the “process_c3ds_healthy.v3s” pipeline can be used instead of the pipeline used for the stroke survivors. The model information needed is stored in the calibration file and not the dynamic file, finally the subject IDs are not consistent throughout all the files so a new naming convention must be made for the CMZ files.
Edit the pipeline command “Apply_Model_Template” in the pipeline process_c3ds_healthy.v3s to point to the model file you have built (or been provided: healthy_model.mdh).
Run the pipeline process_c3ds_healthy.v3s, and select the folder containing the able-bodied participants (138_HealthyPiG_10.05) when prompted.
Set a pipeline parameter to 0, this will be used for the naming of the CMZ file. The value will be iterated within the loop:
Set_Pipeline_Parameter /PARAMETER_NAME=SUB_NUM /PARAMETER_VALUE=0 ! /PARAMETER_VALUE_SEARCH_FOR= ! /PARAMETER_VALUE_REPLACE_WITH= ! /PARAMETER_VALUE_PREFIX= ! /PARAMETER_VALUE_APPEND= ! /MULTI_PASS=FALSE ;
Set a pipeline parameter to a list of folders containing C3Ds within the 138_Healthy_PiG_10.05 folder:
Set_Pipeline_Parameter_To_Folder_Path /PARAMETER_NAME=FOLDER ! /PARAMETER_VALUE=VISUAL3D_DEFAULT_DATA_FOLDER ! /FROM_MOTION_FILE_IN_WORKSPACE= ! /PARAMETER_VALUE_SEARCH_FOR= ! /PARAMETER_VALUE_REPLACE_WITH= ! /PARAMETER_VALUE_APPEND= ! /SET_PROMPT=Select a directory ! /ALPHABETIZE=TRUE ; Set_Pipeline_Parameter_To_List_Of_Files /PARAMETER_NAME=SUB_FOLDERS /FOLDER=::FOLDER /SEARCH_SUBFOLDERS=TRUE /FILE_MASK=*.c3d ! /ALPHABETIZE=TRUE /RETURN_FOLDER_NAMES=TRUE ;
Loop through the folders:
For_Each /ITERATION_PARAMETER_NAME=FOLDER ! /ITERATION_PARAMETER_COUNT_NAME= /ITEMS=::FOLDERS ;
Calibrations files are numbered (0) so get a list of all files ending with 0) (There should only be one in each folder):
Set_Pipeline_Parameter_To_List_Of_Files /PARAMETER_NAME=Cal_File /FOLDER=::FOLDER ! /SEARCH_SUBFOLDERS=FALSE /FILE_MASK=*0)*.c3d ! /ALPHABETIZE=TRUE ! /RETURN_FOLDER_NAMES=FALSE ;
Open a new workspace:
File_New ;
Open the calibration file:
File_Open /FILE_NAME=::CAL_FILE ! /FILE_PATH= ! /SUFFIX= ! /SET_PROMPT=File_Open /ON_FILE_NOT_FOUND=FAIL ! /FILE_TYPES_ON_PROMPT= ;
Set the calibration file to active:
Select_Active_File /FILE_NAME=::CAL_FILE ! /QUERY= ! /SUBJECT_TAGS=NO_SUBJECT ;
Save all the necessary information stored in PARAMETERS:PROCESSING into pipeline parameters:
Set_Pipeline_Parameter_To_Data_Value /SIGNAL_TYPES= PARAMETERS /SIGNAL_FOLDER= PROCESSING /SIGNAL_NAMES= Bodymass ! /SIGNAL_COMPONENTS=ALL_COMPONENTS /PARAMETER_NAME= BODYMASS ; ... See attached pipeline for full list of commands ...
Visual 3Ds expected unit of measurement is meters so the knee widths, ankle widths and height need to be converted from mm to m:
Set_Pipeline_Parameter_From_Expression /PARAMETER_NAME=R_KNEE_WIDTH /EXPRESSION=&::R_KNEE_WIDTH&/1000.0 /AS_INTEGER=False ; ... See attached pipeline for full list of commands ...
Close the calibration file:
File_Close /FILE_NAME=::CAL_FILE ! /QUERY= ! /CLOSE_ASSOCIATED_MODELS=FALSE ;
To open the dynamic files without re-opening the calibration file, list all C3D files within the current folder:
Set_Pipeline_Parameter_To_List_Of_Files /PARAMETER_NAME=DYNAMIC_FILES /FOLDER=::FOLDER ! /SEARCH_SUBFOLDERS=FALSE /FILE_MASK=*.c3d ! /ALPHABETIZE=TRUE ! /RETURN_FOLDER_NAMES=FALSE ;
Loop through each file:
For_Each /ITERATION_PARAMETER_NAME=DYNAMIC_FILE ! /ITERATION_PARAMETER_COUNT_NAME= /ITEMS=::DYNAMIC_FILES ;
Use a conditional statement to check that the current file name does not match the calibrations file name:
Conditional_Statement /ITERATION_PARAMETER_NAME=IS_DYNAMIC /EXPRESSION= "&::DYNAMIC_FILE&" >< "&::CAL_FILE&" ! /AS_INTEGER=TRUE ;
if the file names do not match open the selected file:
File_Open /FILE_NAME=::DYNAMIC_FILE ! /FILE_PATH= ! /SUFFIX= ! /SET_PROMPT=File_Open ! /ON_FILE_NOT_FOUND=PROMPT ! /FILE_TYPES_ON_PROMPT= ;
Close the conditional statement and the loop
Conditional_Statement_End /ITERATION_PARAMETER_NAME=IS_DYNAMIC ; End_For_Each /ITERATION_PARAMETER_NAME=DYNAMIC_FILE ;
Set all dynamic files to active:
Select_Active_File /FILE_NAME=::FOLDER&SUBJ*.c3d ! /QUERY= ! /SUBJECT_TAGS=NO_SUBJECT ;
Create a hybrid model using the calibration file:
Create_Hybrid_Model /CALIBRATION_FILE=::CAL_FILE ! /RANGE=ALL_FRAMES ;
Apply the model template:
/CALIBRATION_FILE=::CAL_FILE !/SUBJECT_PREFIX= /MODEL_TEMPLATE=!!! SET PATH TO HEALTHY MODEL TEMPLATE !!! ! /SET_PROMPT=Open model file ! /VIEW_BUILDMODEL_RESULTS=2 ! /MISSING_TARGET_MESSAGE=FALSE ;
Set the necessary model metrics using the parameters saved from the calibration file and then recalculate the model:
Set_Model_Metric ! /CALIBRATION_FILE= /METRIC_NAME= Mass /METRIC_VALUE= ::BODYMASS ! /SUBJECT= ; ... See the attached pipeline for full list of commands ...
Assign model to the dynamic files:
Assign_Model_File /CALIBRATION_FILE=::CAL_FILE /MOTION_FILE_NAMES=::FOLDER&SUBJ*.c3d ! /REMOVE_EXISTING_ASSIGNMENTS=FALSE ;
Run all desired computations, and then iterate the SUB_NUM pipeline parameter:
... See attached pipeline for examples of computation commands ... Set_Pipeline_Parameter_From_Expression /PARAMETER_NAME=SUB_NUM /EXPRESSION=&::SUB_NUM&+1 ! /AS_INTEGER=TRUE ;
Save the workspace as a CMZ appending the SUB_NUM variable to the name, then close the loop:
/FILE_NAME=SUBJ&::SUB_NUM&.cmz /FOLDER=::FOLDER ! /SET_PROMPT=Save CMZ file as ! /SAVE_EMBEDDED_GRAPHICS=FALSE ! /CREATE_FOLDER_PATH=FALSE ; End_For_Each /ITERATION_PARAMETER_NAME= FOLDER ;
Missing Parameters
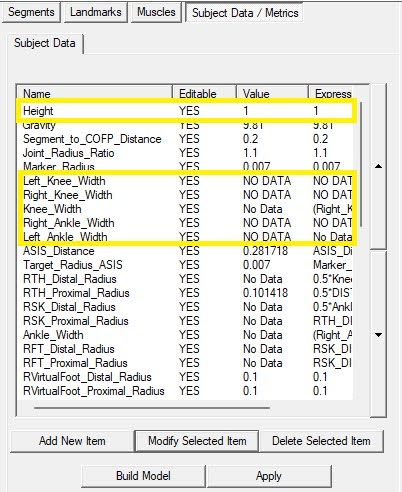
Upon inspection some of the dynamic files were missing or have incorrectly named parameters needed to calculate the model (height, mass, ankle widths, and knee widths) in the case of the stroke participants this data was provided in a spreadsheet so the values can be entered manually and the models recalculated.
Open the CMZ one of Stroke Survivors 24, 34, or 57 (TVC_24.cmz, TVC_34.cmz, and TVC_57.cmz) . Edit the “Subject Data/Metrics” Height, Mass, Left_Knee_Width, Right_Knee_Width, Left_Ankle_Width, and Right_Ankle_Width to match those in the file SubjectChar.xlsx.
Run the pipeline pipeline_manual.v3s while the CMZ is open to recalculate the desired metrics.
Do this separately for all 3 stroke survivors.
After completing this, you will have a completely processed dataset! You can follow other tutorials to see what analysis you might do with this data afterwards.
Conclusion
Through this tutorial we have identified the common pitfalls that may occur when processing a large biomechanics dataset. Using the public dataset from Van Criekinge et al. as an example, we have identified the challenges encountered and presented realistic solutions. Just like the underlying data set, we made all of our files openly available. You are encouraged to work with this dataset as we did and to build upon our work by analyzing the post-processed data.
References
Paper: Van Criekinge et al. a full-body motion capture gait dataset of 138 able-bodied adults across the life span and 50 stroke survivors: [2]