EMG: Difference between revisions
| (26 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
== Overview == | == Overview == | ||
Electromyography (EMG) signals are electrical signals generated by the activity of skeletal muscles | Electromyography (EMG) signals are electrical signals generated by the activity of skeletal muscles when they respond to inputs from the central nervous system. When a muscle contracts, it generates electrical activity that can be measured using electrodes. The electrodes can be placed on the surface of the skin or they can be intramuscular (inserted under the skin with a narrow needle). EMG data is collected as a voltage difference between a recording site and a reference site. Typically the recording site will be the fleshy part of a muscle and the reference site would be near a bony part of the body. EMG signals are used as diagnostics tools for identifying neuromuscular disorders or in research for studying muscle activation, motor control and muscle fatigue. They can also be used as control signals for prosthetic devices. | ||
== Sources of Noise in EMG Data == | == Sources of Noise in EMG Data == | ||
One of the challenges with collecting EMG data is that because the signals are very small (typically | [[File:ArmElectrodePlacement.jpg|right|300 px]] | ||
One of the challenges with collecting EMG data is that because the signals are very small (typically in the range of 0 - 10 mV), and the electrodes used to measure them are prone to picking up other signals that can obscure the data researchers are interested in collecting. | |||
Sources of noise in EMG signals include: | Sources of noise in EMG signals include: | ||
| Line 33: | Line 34: | ||
:*Technical issues with data collection (e.g. the saved EMG signal might be "delayed" because the EMG system is wireless) | :*Technical issues with data collection (e.g. the saved EMG signal might be "delayed" because the EMG system is wireless) | ||
You must know the sync between the analog and motion capture data, | You must know the sync between the analog and motion capture data, and you can use the [[Shift_Frames]] command to synchronize the analog data to the motion capture data. | ||
== Further Information == | |||
'''[[EMG Processing Tools |Processing Tools]]:''' High level descriptions of Visual3D features for general processing of EMG data, with links to detailed examples. | |||
'''Feature Extraction:''' Visual3D pipeline commands for extracting statistical features in EMG applications (in development). | |||
'''[[EMG Detecting Muscle Onset | Detecting Muscle Onset]]:''' Examples of using Visual3D pipeline commands to detect the onset of muscle activation, with examples. | |||
'''[[Tutorial EMG | Tutorial: EMG Processing Workflow]]:''' Basic start to finish tutorial. | |||
== References == | == References == | ||
| Line 43: | Line 54: | ||
3. [http://www.seniam.org/ Surface Electromyography For Non-Invasive Assessment of Muscles] | 3. [http://www.seniam.org/ Surface Electromyography For Non-Invasive Assessment of Muscles] | ||
4. [http://www.humankinetics.com/products/all-products/research-methods-in-biomechanics-2nd-edition Robertson G, Caldwell G, Hamill J, Kamen G, Whittlesey S. Research Methods in Biomechanics | 4. [http://www.humankinetics.com/products/all-products/research-methods-in-biomechanics-2nd-edition Robertson G, Caldwell G, Hamill J, Kamen G, Whittlesey S. Research Methods in Biomechanics 2nd Edition] | ||
5. [https://www.mdpi.com/1424-8220/13/9/12431 Surface Electromyography Signal Processing and Classification Techniques] | 5. [https://www.mdpi.com/1424-8220/13/9/12431 Surface Electromyography Signal Processing and Classification Techniques] | ||
6. [https://www.noraxon.com/higher-education/ Noraxon Higher Education] | 6. [https://www.noraxon.com/higher-education/ Noraxon Higher Education] | ||
[[Category:EMG]] | [[Category:EMG]] | ||
Latest revision as of 17:31, 12 May 2023
| Language: | English • français • italiano • português • español |
|---|
Overview
Electromyography (EMG) signals are electrical signals generated by the activity of skeletal muscles when they respond to inputs from the central nervous system. When a muscle contracts, it generates electrical activity that can be measured using electrodes. The electrodes can be placed on the surface of the skin or they can be intramuscular (inserted under the skin with a narrow needle). EMG data is collected as a voltage difference between a recording site and a reference site. Typically the recording site will be the fleshy part of a muscle and the reference site would be near a bony part of the body. EMG signals are used as diagnostics tools for identifying neuromuscular disorders or in research for studying muscle activation, motor control and muscle fatigue. They can also be used as control signals for prosthetic devices.
Sources of Noise in EMG Data
One of the challenges with collecting EMG data is that because the signals are very small (typically in the range of 0 - 10 mV), and the electrodes used to measure them are prone to picking up other signals that can obscure the data researchers are interested in collecting.
Sources of noise in EMG signals include:
- Physiological features: fat, blood flow velocity, skin temperature and different measuring sites can introduce noise into EMG signals.
- Inherent noise in electrodes: electrical equipment generates electrical noise, which can be picked up in EMG measurements.
- Motion artifacts: the motion of cables during EMG measurements can produce noise in EMG data.
- Cross talk: depending on the placement of electrodes relative to the location of the muscle that is being studied, sometimes an EMG can pick up signals from a muscle that is not the intended target of study.
- Signal instability: EMG signals are quasi random in nature. The number of active motor units, motor firing rate and interaction between muscle fibers can influence an EMG signal.
- Electromagnetic noise: ambient noise from the human body and background radiation can obscure the desired signal to be measured.
What to Look For
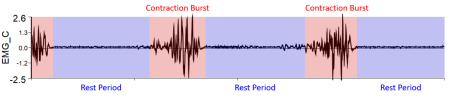
The figure below shows what EMG data should look like. There should be a clear burst (red) when the muscle contracts and periods of rest (blue) when the muscle relaxes. Be sure to check your data for clipping, excessive noise, and movement artifacts, as these cannot be corrected after collection.
Sampling Rate and Synchronization
Sampling Rate
Visual3D expects the EMG signals to be stored as ANALOG data, not unlike force platform data. One point of importance is that EMG typically has very high frequency content, which means that it must be sampled at high data sampling rates.
Synchronization
If you plan to analyze your EMG and motion capture data together, the data must be synchronized. Data can be out of sync due to issues such as:
- A physiological issue known as electromechanical delay (EMD)
- Technical issues with data collection (e.g. the saved EMG signal might be "delayed" because the EMG system is wireless)
You must know the sync between the analog and motion capture data, and you can use the Shift_Frames command to synchronize the analog data to the motion capture data.
Further Information
Processing Tools: High level descriptions of Visual3D features for general processing of EMG data, with links to detailed examples.
Feature Extraction: Visual3D pipeline commands for extracting statistical features in EMG applications (in development).
Detecting Muscle Onset: Examples of using Visual3D pipeline commands to detect the onset of muscle activation, with examples.
Tutorial: EMG Processing Workflow: Basic start to finish tutorial.
References
2. ISEK EMG Reporting Standards
3. Surface Electromyography For Non-Invasive Assessment of Muscles
5. Surface Electromyography Signal Processing and Classification Techniques