XManager: How to Create a Subject: Difference between revisions
m (Wikisysop moved page XManager HowTo Create Subject to XManager: How to Create a Subject without leaving a redirect: page rename) |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 70: | Line 70: | ||
#<i>Surface File</i>: the polygonal file (OBJ) representing the surface of the object, defined in the CT reference frame. If you plan to generate the surface model in [[Surface3D_Overview|Surface3D]], you don't have to specify a file here. However, if you use a third-party program to generate it, you'll have to specify its file name. | #<i>Surface File</i>: the polygonal file (OBJ) representing the surface of the object, defined in the CT reference frame. If you plan to generate the surface model in [[Surface3D_Overview|Surface3D]], you don't have to specify a file here. However, if you use a third-party program to generate it, you'll have to specify its file name. | ||
#<i>ROI Surface File</i>: the same surface model of the object, but with regions of interest and defined in the object's local anatomical reference frame. This file will be generated by [[Orient3D_Overview|Orient3D]]. | #<i>ROI Surface File</i>: the same surface model of the object, but with regions of interest and defined in the object's local anatomical reference frame. This file will be generated by [[Orient3D_Overview|Orient3D]]. | ||
#<i>Inner Surface File</i>: the polygonal file (OBJ) representing the inner boundary of the cortical bone of an object, defined in the CT reference frame. This file is used when tracking bones derived from MRI data. If you create a Surface file representing the outer boundary of the cortical bone, and an Inner Surface File representing the inner boundary, you can create simulated CT data from them using the | #<i>Inner Surface File</i>: the polygonal file (OBJ) representing the inner boundary of the cortical bone of an object, defined in the CT reference frame. This file is used when tracking bones derived from MRI data. If you create a Surface file representing the outer boundary of the cortical bone, and an Inner Surface File representing the inner boundary, you can create simulated CT data from them using the [[Orient3D:_Image_Data_Generator|Image Data Generator]] widget in Orient3D. | ||
[[File:xManFileData.png|800px]] | [[File:xManFileData.png|800px]] | ||
| Line 151: | Line 151: | ||
44) Set the trial name. Choose whether the trial type is <i>reference</i> or <i>motion</i>. Reference trials are usually short, static trials that are used to define a reference pose for multiple motion trials. Motion trials are all other trials (usually ones in which the subject is in motion). | 44) Set the trial name. Choose whether the trial type is <i>reference</i> or <i>motion</i>. Reference trials are usually short, static trials that are used to define a reference pose for multiple motion trials. Motion trials are all other trials (usually ones in which the subject is in motion). | ||
45) If the type is <i>reference</i> and there is motion capture data for this trial, specify the Visual3D model and script files and define the generalized coordinates. See the [[ | 45) If the type is <i>reference</i> and there is motion capture data for this trial, specify the Visual3D model and script files and define the generalized coordinates. See the [[X4D:_How_to_Calculate_Bone_Poses_From_Mocap_Data|Motion Capture Data]] tutorial for details on these parameters. | ||
46) If the type is <i>motion</i>, specify the corresponding C3D file, if any. Also choose the reference trial for this trial. For both types of trials, specify the <i>Time Offset</i>, which is the start time of the X-ray data collection minus the start time of the motion capture data collection. | 46) If the type is <i>motion</i>, specify the corresponding C3D file, if any. Also choose the reference trial for this trial. For both types of trials, specify the <i>Time Offset</i>, which is the start time of the X-ray data collection minus the start time of the motion capture data collection. | ||
Latest revision as of 20:21, 18 January 2021
| Language: | English • français • italiano • português • español |
|---|
1) Select New Subject from the File menu. Use the file browser to create a new .dsx file to hold this subject's information.
2) Click on the default ID (0000) in the tree view in the left panel. The right panel will display the subject information.
3) Specify the ID, subject name, and study description. Time Precision is the number of digits after the decimal point for all time stamps in DSX. This is the precision used to store times in the subject file and to compare times to each other. For Units, it is recommended that you leave it at the default of millimeters.
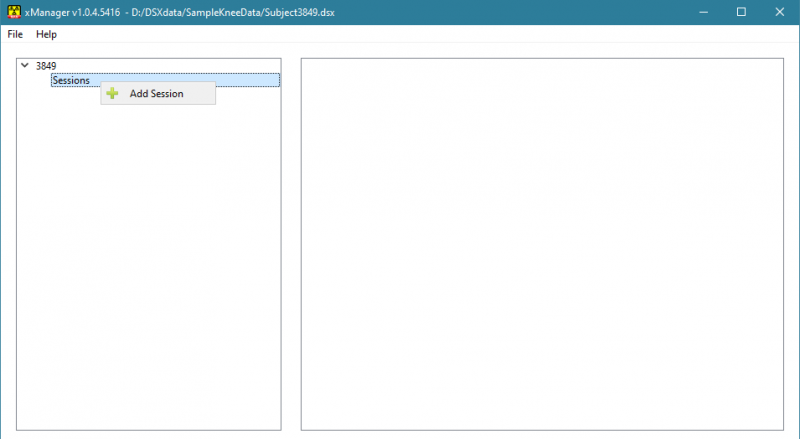
4) Right-click on Sessions in the tree view and select Add Session from the menu.
5) Click on the default name of "newsession" to display the session information in the right panel.
6) Specify the session name and subject metrics.
7) Click on the Paths checkbox. Paths are shortcuts that are used to make data file locations relative to the path value. When a new file path is added to the subject file, xManager will replace part of the file path with a path shortcut if it can find a matching one. The DSX applications use these path shortcuts to keep the data directory structure organized. If you define path shortcuts for all of the data folders for a subject, and make all of them relative to the subject file, you will be able to move an entire subject data folder to a new location on your computer (or another computer) and not have to change any file paths.
8) Right-click on the empty part of the path list and select Add Path from the menu.
9) Click on the browse button on the right to browse for a folder.
10) Double-click on the path's name and enter an appropriate name.
11) Click on the Relative checkbox if you want this shortcut path to be relative to the folder containing the subject file.
12) Repeat steps 8 - 11 for all paths you want to define. You might want one for the X-ray files, one for the CT files, one for the surface model files, and one for the motion capture files. Or you might want one for each trial and/or each X-ray configuration.
13) You do not need to specify the CMO File Name. You will be prompted for a name in one of the other applications when this file is created.
14) Click on the Scan Data checkbox to display the scan data information.
15) Right-click on the empty part of the list and select Add Scan Data from the menu.
16) Click on the edit icon on the right to display the scan data dialog. Use it to select the 3D image data. If the data is a set of 2D images, choose the first image in the file browser.
17) Repeat steps 15 - 16 for all 3D image data sets for this session.
18) Click on the arrow next to the session name in the tree to reveal Trackable Objects, Calibration Devices, Grid Devices, and Configurations. Trackable Objects are the bones or implants that you will be tracking in the X-ray images. Calibration Devices are objects with embedded beads and are used to calculate the 3D configuration of the X-ray equipment. Grid Devices are sheets with a matrix of beads or holes and are used for distortion correction. Configurations are the different configurations of the X-ray equipment during data collection.
19) Right-click on Trackable Objects and select Add Trackable Object from the menu.
20) Click on "newobject" in the tree to show the object information in the panel on the right.
21) Fill in the object information. The Mocap Segment Name is the name of the segment in the Visual3D kinematic model that corresponds to this object. You do not need to specify the Weighted Center; this will be calculated automatically when the object's image data is loaded into X4D.
22) Color is the color of the object's DRR in X4D. It is a good idea to make each object a unique color that is relatively bright so that it shows up well over the white X-ray images.
23) File Data for an object contains:
- Image File: the segmented 3D image data (CT) representing the object. If use use Surface3D to segment the object from the image data, you do not need to specify the file now. If you create it using a third-party program such as Mimics or ScanIP, specify the file here.
- Surface File: the polygonal file (OBJ) representing the surface of the object, defined in the CT reference frame. If you plan to generate the surface model in Surface3D, you don't have to specify a file here. However, if you use a third-party program to generate it, you'll have to specify its file name.
- ROI Surface File: the same surface model of the object, but with regions of interest and defined in the object's local anatomical reference frame. This file will be generated by Orient3D.
- Inner Surface File: the polygonal file (OBJ) representing the inner boundary of the cortical bone of an object, defined in the CT reference frame. This file is used when tracking bones derived from MRI data. If you create a Surface file representing the outer boundary of the cortical bone, and an Inner Surface File representing the inner boundary, you can create simulated CT data from them using the Image Data Generator widget in Orient3D.
24) Reference Frame is the 4x4 transform from the object's CT frame to its local, anatomical reference frame. If you will be computing this frame in Orient3D, you do not need to specify it now.
25) Landmarks are landmarks created in the 3D image data by Surface3D or on the surface model by Orient3D.
26) Points of Interest are points on the object that can be tracked in the X-ray images, such as beads implanted in bones or beads in a calibration object. To add a POI, right-click on the empty part of the POI list and select Add POI from the menu. For the calibration object, create a POI for each bead in the object. The Radius should be the radius of the physical bead, which is usually specified in millimeters.
27) Repeat steps 19 - 26 for all objects in this session.
28) Right-click on Calibration Devices and select Add Calibration Device from the menu.
29) Click on "newcal" in the tree to show the calibration device information in the panel on the right.
Fill in the data for device's points of interest and motion capture markers. The points of interest are the beads embedded in the physical object. For each bead, specify its 3D location and its radius. Specifying a color is optional, but can be helpful when digitizing the bead locations in the X-ray images. The X-ray lab reference frame is implicitly defined by the 3D coordinates of the calibration device's points of interest.
If you want to synchronize your dynamic X-ray images with motion capture data, you need to put at least three motion capture markers on the calibration device and specify their locations and radii. The coordinates of these mocap markers are specified in the X-ray lab reference frame. Once they are tracked by the motion capture system during collection of the X-ray images of the calibration device, the transform from the X-ray lab frame to the motion capture frame can be computed by CalibrateDSX.
30) Right-click on Grid Devices and select Add Grid Device from the menu.
31) Click on "newgrid" in the tree to show the calibration device information in the panel on the right.
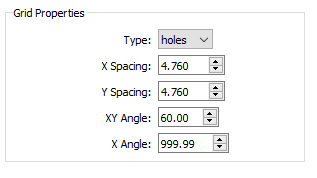
Specify the grid properties.
- Type - the type of grid. Holes refers to a radiopaque sheet with punched or drilled holes. Beads refers to a radiolucent sheet with embedded radiopaque beads.
- X Spacing - the distance, in millimeters, between adjacent beads/holes in the grid object, along the X axis. The default value is 10.0.
- Y Spacing - the distance, in millimeters, between adjacent beads/holes in the grid object, along the Y axis. The default value is 10.0.
- XY Angle - the angle between the X and Y axes of beads/holes in the grid object. For hex grids, this parameter can be set to 60.0 or 120.0. The default value is 90.0.
- X Angle - the angle between the X axis of grid beads/holes and the X axis of the grid image. If the grid X axis is supposed to be aligned with the image intensifiers, set this parameter to 0.0. If the alignment of the grid X axis is unknown (common with hex grids of holes), set this parameter to a value outside the range -360.0 to 360.0, which tells CalibrateDSX to calculate the alignment. The default value is 999.9.
32) Right-click on Configurations and select Add Configuration from the menu.
33) Click on "newconfig" in the tree to display the configuration information in the panel on the right.
34) Set the name and date. The Lab to Mocap Transformation will be calculated by CalibrateDSX when you calibrate the system.
35) Click on the arrow next to the configuration name in the tree to reveal Setup, Calibration, and Trials.
36) Click on the arrow next to Setup to reveal Views, then click on the arrow next to Views to reveal the two default X-ray views.
37) Click on each view and set its name. The other parameters are calculated in CalibrateDSX when you compute the 3D X-ray configuration.
38) Click on Calibration in the tree to reveal the source-to-image distances for the two views in the right panel. Enter these distances, in millimeters, as measured on the physical X-ray components (X-ray source to plane containing the grid object). It is important to measure these distance accurately, as the calculation of the 3D configuration is fairly sensitive to their values.
39) Click on the arrow next to Calibration to reveal Trials, then click on the arrow next to Trials to reveal the three [required] calibration trials.
40) For each of the three calibration trials:
- Click on it in the tree to display its information in the panel on the right.
- Click on the Views checkbox to reveal the views.
- Click on the first view in the list. Specify the Raw File containing the uncorrected X-ray data for this view.
- Click on the second view in the list. Specify the Raw File containing the uncorrected X-ray data for this view.
41) For the Distortion trial, select the grid device that you created in steps 30 and 31.
42) Right-click on Trials (not Calibration->Trial) in the tree and select Add Trial from the menu.
43) Click on "newtrial" in the tree to display the trial information in the panel on the right.
44) Set the trial name. Choose whether the trial type is reference or motion. Reference trials are usually short, static trials that are used to define a reference pose for multiple motion trials. Motion trials are all other trials (usually ones in which the subject is in motion).
45) If the type is reference and there is motion capture data for this trial, specify the Visual3D model and script files and define the generalized coordinates. See the Motion Capture Data tutorial for details on these parameters.
46) If the type is motion, specify the corresponding C3D file, if any. Also choose the reference trial for this trial. For both types of trials, specify the Time Offset, which is the start time of the X-ray data collection minus the start time of the motion capture data collection.
47) Click on Tracked Bodies to reveal the list of objects that are defined in the Objects section of the tree. For each of the objects in the list that can be tracked in this trial, check its box. If you click on the object name, the object's data files will be shown on the right. All of these files will be generated later, when the object is tracked in the X-ray images.
48) Click on the Views checkbox at the bottom of the right panel to reveal the views.
49) Click on the first view in the list. Specify the Raw File containing the X-ray data for this view.
50) Click on the second view in the list. Specify the Raw File containing the X-ray data for this view.
51) Repeat steps 37 - 45 for all trials in this configuration.
52) Repeat steps 28 - 46 for all configurations in this session.
53) Repeat steps 4 - 47 for all sessions in this subject.